Here's some tricks that are allowed by pandoc but not obvious at first sight.
Here's some tricks that are allowed by pandoc but not obvious at first sight.
-----
**Table of Contents**
<!-- MarkdownTOC autolink="true" bracket="round" autoanchor="false" lowercase="true" lowercase_only_ascii="true" uri_encoding="true" depth="3" -->
- [From Markdown, To Markdown](#from-markdown-to-markdown)
- [Cleanup](#cleanup)
- [TOC generation](#toc-generation)
- [Using Markdown Templates](#using-markdown-templates)
- [Math in Pure Markdown](#math-in-pure-markdown)
- [Convert Between the 4 Table Syntaxes in Pandoc](#convert-between-the-4-table-syntaxes-in-pandoc)
- [Repeated Footnotes Anchors and Headers Across Multiple Files](#repeated-footnotes-anchors-and-headers-across-multiple-files)
- [Template Snippet](#template-snippet)
- [YAML Metadata for Any Format](#yaml-metadata-for-any-format)
- [Left-aligning Tables in LaTeX](#left-aligning-tables-in-latex)
- [GFM Task Lists with Pandoc](#gfm-task-lists-with-pandoc)
- [Via Lua Filter](#via-lua-filter)
- [Via PP Macros](#via-pp-macros)
- [Today in date metadata](#today-in-date-metadata)
<!-- /MarkdownTOC -->
-----
# From Markdown, To Markdown
using `pandoc -f markdown... -t markdown...` can have surprisingly useful applications. As a demo, this file is generated by
``` bash
pandoc -t gfm --atx-headers \
--reference-location=block --toc -s -o temp-github.md temp.md
```
Be careful of `@` though, you need to escape it in pandoc since it is treated as citation in pandoc.
## Cleanup
As shown in issue [\#2814](https://github.com/jgm/pandoc/issues/2814), rendering a document to itself can be used to clean up / normalize your markdown file.
## TOC generation
*e.g.* you have a long markdown file in GitHub and want to have a TOC, you can use `pandoc -t gfm --toc -o example-with-toc.md example.md`
This a useful workaround to update the TOC of very long documents, but —beware!— if you use this trick for writing over the input file, you'll end stacking TOCs — each new Table of Contents being generated above the previously built ones, and indexing them too. This technique is useful when working with different source and output files.
Also, you can add a title to the TOC using the `toc-title` variable, but only if you use a markdown template — as explained ahead.
## Using Markdown Templates
Did you know that you can use pandoc template with markdown too?
Ask pandoc to write-out the default template for markdown:
``` bash
pandoc --print-default-template=markdown > template.markdown
```
And now let's peek at the template we got:
```
$if(titleblock)$
$titleblock$
$endif$
$for(header-includes)$
$header-includes$
$endfor$
$for(include-before)$
$include-before$
$endfor$
$if(toc)$
$toc$
$endif$
$body$
$for(include-after)$
$include-after$
$endfor$
```
As you can see, there's plenty of conditional statements to play with, allowing for additional control over the output markdown file.
You can also use the `toc-title` template variable to tell pandoc to add a title on top of the generated TOC. Change the template’s `toc` block like this:
```
$if(toc)$
$if(toc-title)$
# $toc-title$
$endif$
$toc$
$endif$
```
And now invoke pandoc like this:
``` bash
pandoc --toc -V toc-title:"Table of Contents" --template=template.markdown -o example-with-toc.md example.md
```
And you'll see in the `example-with-toc.md` file an auto-generated Table of Contents with a `# Table of Contents` title over it.
> **NOTE**: if you also include some extra markdown contents with the `--include-before-body` option (eg: `--include-before-body=somefile.md`) the contents of the included file will go before the TOC (at least, with the template used in this example) and any headings it contains will not be included in the TOC — ie: the TOC only indexes what comes after the `$toc$` template tag. This is useful if you'd like to include an Abstract before the TOC.
## Math in Pure Markdown
The manual said:
> Note: the `--webtex` option will affect Markdown output as well as HTML.
This can be used to put math in pure markdown. *e.g.* you want to put math directly in the README.md in GitHub.
For example, in the `temp.md`:
``` markdown
# Important Discovery!
$1+2\neq3!$
Try it!
```
Run this:
``` bash
pandoc --atx-headers --webtex=https://latex.codecogs.com/png.latex? -s -o temp-codecogs.md temp.md
```
Then the output becomes:
> # Important Discovery!
>
> 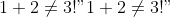
>
> Try it!
## Convert Between the 4 Table Syntaxes in Pandoc
Say, in your source markdown file `pipe.md`:
``` markdown
| testing | pandoc | tables |
|-------------|-------------------|---------|
| simple cell | no multiline cell | and |
| so | on | no list |
```
In command line,
``` bash
pandoc -t markdown-simple_tables-multiline_tables-pipe_tables -s -o grid.md pipe.md
```
In the output `grid.md`:
``` markdown
+--------------------------+--------------------------+--------------------------+
| testing | pandoc | tables |
+==========================+==========================+==========================+
| simple cell | no multiline cell | and |
+--------------------------+--------------------------+--------------------------+
| so | on | no list |
+--------------------------+--------------------------+--------------------------+
```
## Repeated Footnotes Anchors and Headers Across Multiple Files
If you use auto-identifiers for the headers, and there are different headers with the same name across different files, you'd want to catenate them together, and pandoc can do this for you:
``` bash
pandoc file1.md file2.md ...
```
But if there are repeated footnotes anchors on both files, you need to use the `--file-scope` option, which will parse each file individually (so the footnotes anchors are "local" to the individual file):
``` bash
pandoc file1.md file2.md --file-scope ...
```
What about if the 2 files have both these problems? i.e., headers with same names (hence the same Id by the auto-identifier) and footnotes with same anchors appear across the files. Either approach gives you problems.
In this case, you can use "to markdown from markdown" to write an intermediate markdown file using `--file-scope`, which handles the colliding footnote anchors for you, and then generate the final document from that intermediate markdown file, and let the auto-identifiers handle the headers for you:
``` bash
pandoc --file-scope -o intermediate.md file1.md file2.md
pandoc intermediate.md ...
```
# Template Snippet
If you wrote a template snippet that do not form a complete template. The `-H`, `-B`, or `-A` option would not help because pandoc would put your snippet as is and wouldn't process it as a template. *i.e.* The snippet is included after the template is processed.
A trick mentioned by [@cagix](https://github.com/cagix) in [jgm/pandoc-templates\#220](https://github.com/jgm/pandoc-templates/issues/220) is this:
``` bash
pandoc --template=template_snippet.tex document.md -o processed_snippet
pandoc ... -H processed_snippet document.md -o document.<toFormat>
# Or shorter but bash only (process substitution)
SNIPPET=template_snippet.tex; INPUT=document.md; OUTPUT=document.<toFormat>
pandoc ... -H <(pandoc --template=$SNIPPET $INPUT) $INPUT -o $OUTPUT
```
The first line will process your template snippet according to the properties of the document, but since your snippet (probably) do not have `$body$`, the body would not be in the output. Now the snippet is processed and can then be included through `-H` as is in the 2nd line.
# YAML Metadata for Any Format
YAML metadata is only defined for pandoc's markdown syntax. See [jgm/pandoc#1960](https://github.com/jgm/pandoc/issues/1960#issuecomment-273587588).
Currently, there is a workaround like this (while the YAML metadata only accepts markdown syntax):
```bash
pandoc -f markdown -t native -s metadata.yml | sed '$ d' > metadata.native
pandoc -t native -o document.native document.<fromFormat>
pandoc -f native -s -o document.<toFormat> metadata.native document.native
# Or shorter but bash only (process substitution)
YAML=metadata.yml; INPUT=document.<fromFormat>; OUTPUT=document.<toFormat>
pandoc ... -f native -s -o $OUTPUT <(pandoc -f markdown -t native -s $YAML | sed '$ d') <(pandoc -t native $INPUT)
```
Explanation:
The `sed` in the first line: because the `metadata.yml` is regarding as a markdown document with no body, so the last line of the metadata in native format is `[]`, which you need to remove. Another way of removing it is `head -n -1` (would not work on Mac's default `head`). From my test it seems the meta in native is always in one-line, if true then `head -n1` will work (which also works on Mac).
# Left-aligning Tables in LaTeX
Based on [this pandoc-discuss exchange](https://groups.google.com/forum/#!topic/pandoc-discuss/pmiw78NvZl4) and [this TeX StackExchange topic](http://tex.stackexchange.com/questions/6456/set-a-global-policy-for-floats-positioning), it is possible to left-align all tables in a document (in the PDF output from LaTeX) with this single invocation in the YAML header block of the markdown document:
`````` yaml
---
header-includes:
- |
```{=latex}
\usepackage[margins=raggedright]{floatrow}
```
...
``````
This applies to all floats, and fine-grained control may be achieved with the options outlined in [the documentation for the `floatrow` LaTeX package](http://mirrors.ctan.org/macros/latex/contrib/floatrow/floatrow.pdf).
# GFM Task Lists with Pandoc
There are currently two ways to allow support of GitHub Flavored Markdown Task Lists:
1. Via the [Task-List Lua] pandoc filter
2. Via custom PP macros
The former is the recommended way, the latter was introduced before the Lua filter came into place.
## Via Lua Filter
Download the [`task-list.lua`][task-list.lua] file and save it in `$DATADIR/pandoc/filters/`, so it will be visible to pandoc system-wide. Now, when you invoke pandoc with the `--lua-filter=task-list.lua`, Task Lists in markdown source documents will be supported as on GitHub.
Command line usage example:
```shell
pandoc --lua-filter=task-list.lua -o filename.html filename.md
```
The Task Lists filter will also take care of injecting the required CSS in HTML documents, in order to represent the Task List styles correctly.
From the [Task-List Lua] documentation:
> This filter recognizes this syntax and converts the ballot boxes into a representation suitable for the chosen output format. Ballot boxes are rendered as
>
> - checkbox input elements in HTML,
> - ASCII boxes in gfm and org-mode, and
> - ballot box UTF-8 characters otherwise.
[Task-List Lua]: https://github.com/pandoc/lua-filters/tree/master/task-list "Visit the pandoc 'lua-filters' repository"
[task-list.lua]: https://github.com/pandoc/lua-filters/blob/master/task-list/task-list.lua "See the source file of 'task-list.lua' on GitHub"
## Via PP Macros
GitHub flavored markdown's task lists can be mimicked in pandoc via the `GFM-TaskList.pp` custom pp macros module. Macro syntax example:
!TaskList(
!Task[x][I'm a _checked_ task]
!Task[ ][I'm an _unchecked_ task]
)
The PP macros approach is more verbose and breaks a document's comptability outside of PP usage context (eg, a `README.md` file on GitHub); this solution was deviced for pandoc v1.x, long before the [Task-List Lua] solution was available. While it might still be usefeul in some specific contexts, using the Lua filter approach is recommended instead.
For more information, and to download the `GFM-TaskList.pp` macros module:
- ["Pandoc-Goodies" project: pp-macros library](https://github.com/tajmone/pandoc-goodies/tree/master/pp)
- [Live HTML preview of Task List created with pandoc and pp macros](https://htmlpreview.github.io/?https://github.com/tajmone/pandoc-goodies/blob/master/templates/html5/github/GitHub-Template-Preview.html#task-lists)
# Today in date metadata
Add this to the pandoc command you use:
```bash
-M date="`date "+%B %e, %Y"`"
```
POSIX only.